Abstract
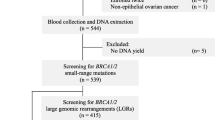
Heritable breast cancers account for 5% to 10% of all breast cancers, and monogenic, highly penetrant genes cause them. Around 90% of pathogenic variants in BRCA1 and BRCA2 are observed using gene sequencing, with another 10% identified through gene duplication/deletion analysis, which differs across various communities. In this study, we performed a next-generation sequencing panel and MLPA on 1484 patients to explain the importance of recurrent germline duplications/deletions of BRCA1-2 and their clinical results and determine how often BRCA gene LGRs were seen in people suspected of hereditary breast and ovarian cancer syndrome. The large genomic rearrangements (LGRs) frequency was approximately 1% (14/1484). All 14 mutations were heterozygous and detected in patients with breast cancer. BRCA1 mutations were more predominant (n = 8, 57.1%) than BRCA2 mutations (6, 42.9%). The most common recurrent mutations were BRCA2 exon three and BRCA1 exon 24 (23) deletions. To the best of our knowledge, BRCA1 5’UTR-exon11 duplication has never been reported before. Testing with MLPA is essential to identify patients at high risk. Our data demonstrate that BRCA1-2 LGRs should be considered when ordering genetic testing for individuals with a personal or family history of cancer, particularly breast cancer. Further research could shed light on BRCA1-2 LGRs’ unique carcinogenesis roles.



Similar content being viewed by others
Data availability
The data (including patient demographic information and mutations) and the code of the current study are available from the corresponding author on reasonable request.
References
Howlader N, Noone A, Krapcho M, et al (2015) SEER Cancer Statistics Review. National Cancer Institute 1975–2012. http://seer.cancer.gov/csr/1975_2012/, based on November 2014 SEER data submission, posted to the SEER web site.
Randall LM, Pothuri B (2016) The genetic prediction of risk for gynecologic cancers. Gynecol Oncol 141:10–16. https://doi.org/10.1016/j.ygyno.2016.03.007
Valencia OM, Samuel SE, Viscusi RK et al (2017) The role of genetic testing in patients with breast cancer a review. JAMA Surg 152:589–594
Eccles BK, Copson E, Maishman T et al (2015) Understanding of BRCA VUS genetic results by breast cancer specialists. BMC Cancer. https://doi.org/10.1186/s12885-015-1934-1
Mavaddat N, Peock S, Frost D et al (2013) Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst 105:812–822. https://doi.org/10.1093/jnci/djt095
Judkins T, Rosenthal E, Arnell C et al (2012) Clinical significance of large rearrangements in BRCA1 and BRCA2. Cancer 118:5210–5216. https://doi.org/10.1002/cncr.27556
Thompson D, Easton DF (2002) Cancer incidence in BRCA1 mutation carriers. J Natl Cancer Inst 94:1358–1365. https://doi.org/10.1093/jnci/94.18.1358
Sluiter MD, Van Rensburg EJ (2011) Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res Treat 125:325–349. https://doi.org/10.1007/s10549-010-0817-z
Preisler-Adams S, Schönbuchner I, Fiebig B et al (2006) Gross rearrangements in BRCA1 but not BRCA2 play a notable role in predisposition to breast and ovarian cancer in high-risk families of German origin. Cancer Genet Cytogenet 168:44–49. https://doi.org/10.1016/j.cancergencyto.2005.07.005
Armour JAL, Barton DE, Cockburn DJ, Taylor GR (2002) The detection of large deletions or duplications in genomic DNA. Hum Mutat 20:325–337
Toland AE, Forman A, Couch FJ et al (2018) Clinical testing of BRCA1 and BRCA2: a worldwide snapshot of technological practices. NPJ Genomic Med. https://doi.org/10.1038/s41525-018-0046-7
Brandt T, Sack LM, Arjona D et al (2020) Adapting ACMG/AMP sequence variant classification guidelines for single-gene copy number variants. Genet Med 22:336–344. https://doi.org/10.1038/s41436-019-0655-2
Vendrell JA, Vilquin P, Larrieux M et al (2018) Benchmarking of amplicon-based next-generation sequencing panels combined with bioinformatics solutions for germline BRCA1 and BRCA2 alteration detection. J Mol Diagnostics 20:754–764. https://doi.org/10.1016/j.jmoldx.2018.06.003
Hall MJ, Reid JE, Burbidge LA et al (2009) BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer 115:2222–2233. https://doi.org/10.1002/cncr.24200
Engert S, Wappenschmidt B, Betz B et al (2008) MLPA screening in the BRCA1 gene from 1,506 German hereditary breast cancer cases: novel deletions, frequent involvement of exon 17, and occurrence in single early-onset cases. Hum Mutat 29:948–958. https://doi.org/10.1002/humu.20723
Pavlicek A, Noskov VN, Kouprina N et al (2004) Evolution of the tumor suppressor BRCA1 locus in primates: implications for cancer predisposition. Hum Mol Genet 13:2737–2751. https://doi.org/10.1093/hmg/ddh301
Caputo SM, Léone M, Damiola F et al (2018) Full in-frame exon 3 skipping of BRCA2 confers high risk of breast and/or ovarian cancer. Oncotarget 9:17334–17348. https://doi.org/10.18632/oncotarget.24671
Bozsik A, Pócza T, Papp J et al (2020) Complex characterization of germline large genomic rearrangements of the BRCA1 and BRCA2 genes in high-risk breast cancer patients—novel variants from a large national center. Int J Mol Sci 21:1–17. https://doi.org/10.3390/ijms21134650
Apostolou P, Pertesi M, Aleporou-Marinou V et al (2017) Haplotype analysis reveals that the recurrent BRCA1 deletion of exons 23 and 24 is a Greek founder mutation. Clin Genet 91:482–487. https://doi.org/10.1111/cge.12824
Muller D, Rouleau E, Schultz I et al (2011) An entire exon 3 germline rearrangement in the BRCA2 gene: pathogenic relevance of exon 3 deletion in breast cancer predisposition. BMC Med Genet. https://doi.org/10.1186/1471-2350-12-121
Armstrong N, Ryder S, Forbes C et al (2019) A systematic review of the international prevalence of BRCA mutation in breast cancer. Clin Epidemiol 11:543–561
Petrucelli N, Daly MB, Pal T (2016) BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer. GeneReviews®. University of Washington, Seattle. https://www.ncbi.nlm.nih.gov/books/NBK1247/
Smith MJ, Urquhart JE, Harkness EF et al (2016) The contribution of whole gene deletions and large rearrangements to the mutation spectrum in inherited tumor predisposing syndromes. Hum Mutat 37:250–256. https://doi.org/10.1002/humu.22938
Funding
No financial assistance was received.
Author information
Authors and Affiliations
Contributions
IS designed the research, analyzed and interpreted the results (including the coding part with Python version 3.9.2), wrote the manuscript, and approved the final manuscript. HS collected the data, analyzed and interpreted the results, reviewed the manuscript, and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical approval
We confirm that we have read the journal’s position on issues involved in the ethical publication, and we affirm that this report is consistent with those guidelines. The Ethics Committee of the University of Health Sciences, Dışkapı Yıldırım Beyazıt Training and Research Hospital approved the study (08.03.2021-106/27).
Informed consent
Informed consent was obtained from the patients or their parents (mentioned within “Patients”).
Consent to participate
Consent for the participation was taken from the patients or their parents involved in the study.
Consent for publication
Consent for the publication was taken from the patients or their parents involved in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sahin, I., Saat, H. A novel BRCA1 duplication and new insights on the spectrum and frequency of germline large genomic rearrangements in BRCA1/BRCA2. Mol Biol Rep 48, 5057–5062 (2021). https://doi.org/10.1007/s11033-021-06499-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06499-3