Abstract
Legionella spp. are important waterborne pathogens that are normally transmitted through aerosols. The present work was conducted to investigate the presence of Legionella spp. and its common species in hospital water supplies. Considering the limitations of culture method, polymerase chain reaction (PCR) assays were developed to detect the gene 16S rRNA irrespective of the bacterial serotype. Four well-established DNA extraction protocols (freeze & thaw and phenol-chloroform as two manual protocols and two commercial kits) were tested and evaluated to release DNA from bacterial cells. A total of 45 samples were collected from seven distinct hospitals’ sites during a period of 10 months. The PCR assay was used to amplify a 654-bp fragment of the 16S rRNA gene. Legionella were detected in 13 samples (28.9%) by all of the methods applied for DNA extraction. Significant differences were noted in the yield of extracted nucleic acids. Legionella were not detected in any of the samples when DNA extraction by freeze & thaw was used. Excluding this method and comparing manual protocol with commercial kits, Kappa coefficient was calculated as 0.619 with p < 0.05. Although no meaningful differences were found between the kits, DNA extraction with Bioneer kit exhibited a higher sensitivity than classical Qiagen. Showerheads and cold-water taps were the most and least contaminated sources with 55.5 and 9 percent positive samples, respectively. Moreover two positive samples were identified for species by DNA sequencing and submitted to the Gene Bank database with accession Nos. FJ480932 and FJ480933. The results obtained showed that despite the advantages of molecular assays in Legionella tracing in environmental sources, the use of optimised DNA extraction methods is critical.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Legionella are thin, gram-negative, obligate aerobic and sporeless rods with complex nutritional requirements. Certain species of Legionella like Legionella pneumophila are often strongly associated with asymptomatic infections (Legionnaires' disease) or produce mild cough, sore throat and fever (Pontiac fever) that goes away by itself in a 2-5 day period. The term "legionellosis" may be used to refer to either Legionnaires' disease or Pontiac fever. However, more than 10 known serotypes are implicated in severe pulmonary nosocomial infections, especially in immunocompromised patients as well as in the elderly and subjects already suffering from pulmonary diseases [1–3]. Indeed, twenty-one species of Legionella are pathogens for humans, especially in patients with chronic pulmonary disease within hospitals [4]. The bacterium can be isolated from aquatic and terrestrial habitats as well as from legionellosis patients [3]. Disease occurs after exposure to aquatic settings that promote bacterial growth-the aquatic environment is somewhat stagnant, the water is warm (25°C–42°C), and the water must be aerosolized so that the bacteria can be inhaled into the lungs. Inhalation or micro aspiration of Legionella from contaminated environmental sources such as hot water systems and cooling towers’ water is the most frequent route of transmission. While transmission has also been reported via nebulizer and showers in contaminated water, the infection is not spread from person to person [3, 5, 6].
Outbreaks of legionellosis have been described in numerous countries throughout the world. In 2007, there were 2716 reported cases, near 8 cases per million in the United States (CDC) [7]. Travel-associated outbreaks are commonly recognized [8]. CDC estimates that between 8,000 and 18,000 people are hospitalized with LD in the United States each year [9]. Hospitals are common habitats for the bacterium, where the bacterial niches are amply found and provide the most likely places for susceptible people to contract the diseases. Outbreaks of legionellosis have been reported from hospital patients in many countries with an incidence range of 0 to 47% [10, 11]. Consequently, national Legionella surveillance programs have been established for regular monitoring of environmental samples in these countries [12, 13]. In Iran however, hospital-acquired Legionnaire's Disease has rarely been reported and environmental surveillance for Legionella in hospital water systems to provide useful data for risk assessment and prevention has never been systematically performed.
The polymerase chain reaction (PCR) is considered the most adaptable and prevalent DNA-based assay technique, which is a highly specific and sensitive alternative method to standardize culture isolation, especially when rapid results are required. This method is especially favorable when the samples contain abundant and diverse micro biota and when fastidious and slow-growing bacteria like Legionella need to be detected. So despite the fact that culture method for isolation of Legionella is the golden standard and has been approved by ISO and many other national standards for water quality determination, over the past few years, molecular techniques based on 16S rRNA gene besides other genetic markers have been developed to analyze bacterial communities in environmental samples [14, 15]. Indeed, considering the complexity of bacterial behaviors which are not easily predictable, PCR can be economically profitable but requires attention in evaluating the suitability and consistency of the used tools and recipes in order to select the best appropriate and efficient ones and consequently, achieving the best results. It has been demonstrated that the application of PCR depends on the extraction of DNA from the organisms, which is often the most critical step to avoid false negative PCR results. In general, the extraction of non-degraded and inhibitor-free DNA, suitable for PCR amplification, has been reported as a common issue [16–18]. Several methods including both commercial kits and manual classic protocols have been used for the preparation of Legionella DNA from environmental samples and evaluation of the quantity and quality of extracted DNA. Several studies have reported that DNA extraction methods can influence the sensitivity of PCR assays [19–22]. Therefore, selection of the best method for a given sample is of importance for the laboratory detection of given organism. This cross-sectional study was conducted from the above perspective to investigate the presence of bacteria belonging to the Legionella genus in water supplies of some hospitals in Tehran, capital city of Iran. The impact of water quality on Legionella existence was also determined. In the present study, four well-established but different methods (freeze & thaw, phenol-chloroform, and DNA extraction using two different commercial kits) were tested and evaluated to release DNA from bacterial cells in order to find a reliable method. In spite of the large amount of data available for the various PCR methods to identify Legionella species in water samples, to our knowledge, this is the first attempt to gather information on common DNA extraction protocols to monitor the presence of this bacterium and there is no systematic comparative study to assess the relative efficiency of these techniques. For further confirmation, two isolates determined as Legionella spp. were randomly sequenced as well.
Materials and methods
Sample collection and preparation
A total of forty-five samples were collected from distinct sites at seven teaching hospitals under the auspices of Tehran University of Medical Sciences from June 2011 to January 2012. Hospital water facilities sampled included tap cold and hot water, showerhead, hot water tank, and cooling tower water. Samples were collected in 1-litre sterile bottles directly from the outlet. Before sampling, a sterile swab was inserted into faucet outlets and rotated against the interior surface two times clockwise and up-and-down two times to dislodge the sediment.
DNA Extraction, PCR assay, Gel electrophoresis, and DNA sequencing
One liter of samples was filtered through 0.22 μm mixed cellulose ester membrane filters (Schleicher & Schuell) in a stainless-steel filter holder with a water aspirator. Each membrane was aseptically scraped, cut into smaller pieces and placed into sterile, screw-capped containers with 10 ml of the original sample. The samples were then sonicated for 5 min (Bandelin Sonorex) at 35 KHz and shaken for 15 min to dislodge bacterial cells from the membranes. The elute was transferred into a 15 mL conical centrifuge tube and centrifuged (2000 g, 20 min) to remove cell debris. Total DNA was extracted from concentrated water samples using two classic manual methods: freeze & thaw and phenol-chloroform as well as two commercial DNA extraction kits: Flexi Gen DNA Kit Qiagen (Hilden, Germany) and Bioneer Accuprep Genomic DNA Kit according to the manufacture's instruction. Table 1 shows the characteristics of these DNA extraction kits. DNA extraction using freeze & thaw method was conducted by placing of 1 mL of each concentrated water sample within 1.5 mL micro tubes and alternating application of freezing the samples in liquid nitrogen and their incubation in water bath in a temperature of 100°C for three or more times. The suspension was then centrifuged again (18000 g, 10 min) and the supernatant was transferred to new micro tubes. Another manual DNA extraction method of phenol-chloroform was done according to Sambrook standard methods [3].
Extracted DNA was stored at -20°C until using PCR. Amplification reactions were preformed according to what has been described earlier by Hsu [14]. The PCR primers LEG 225 (5'-AAGATTAGCCTGCGTCCGAT-3') and LEG 858 (5'-GTCAACTTATCGCGTTTGCT-3') were used to amplify a 650 bp fragment of the 16S rRNA gene of Legionella species. Each 25 μL of reaction contained 20 ng genomic DNA, 1.5 mM MgCl2 (Roche, Germany), 0.2 mM dNTP (Roche, Germany), 20 pM of each primer, and 1u of Tag DNA polymerase (Roche, Germany) in the PCR buffer (Roche, Germany). The cycling conditions were 94ºC for 5 min, followed by 30 cycles at 95ºC (30 sec), 64ºC and 74ºC for 20 sec each, and 1 cycle of 72ºC for 5 min in Thermocycler (Techne, USA). PCR products were loaded onto a 2% agarose gel containing ethidium bromide. Extracted DNA from cultured Legionella was used for positive control. For more confirmation, the PCR products of two Legionella isolates were sequenced at MWG (Ebersberg, Germany; http://www.mwg-biotech.com).
Statistical analysis
Statistical analysis was performed using SPSS (version 9.0; SPSS Inc, Chicago, IL). Quantitative variables were expressed as mean standard deviation when the data was normally distributed, while variables were expressed as median when the data was not in a normal distribution. The results of the DNA extraction tests were reported as qualitative values (positive and negative) and through a comparison of results obtained, the agreement rates were determined by applying the Kappa coefficient.
Results and discussion
Comparison among DNA extraction methods
Several studies have compared DNA extraction methods and reported that their abilities to recover bacterial DNA were different, indicating that no single DNA extraction method is optimal for all bacteria [19, 20]. In this study, four different procedures (two commercial extraction kits and two classic manual protocols) were evaluated and compared to obtain DNA from hospital water samples. DNA was extracted from water samples using these four methods and followed by PCR amplification of bacterial 16S rRNA gene.
Overall, Legionella were detected in 13 samples (28.9%) by all four methods used for DNA extraction. The comparison of these methods regarding their relative ability to extract DNA and detect Legionella from samples bore significant differences according to the results obtained (Table 2). As shown in this table, Legionella were not detected in any of the samples when DNA extraction by freeze & thaw method was performed. Likewise, samples revealed low levels DNA of Legionella (4/45 corresponding to 8.9%) by Qiagen kit. DNA extraction by phenol-chloroform and Bioneer kit, however, revealed the most positive samples for Legionella, i.e., 8 out of 45 (17.7%) and 12 out of 45 samples (26.6%) showed contamination with Legionella, respectively. Better performance of Bioneer kit may be driven from the fact that columned method was applied for DNA extraction of cells (DNA adsorbed to membrane column), though DNA was precipitated by alcohol in the case of Qiagen kit. Four samples were detected positive by Qiagen DNA extraction kit, which were positive with Bioneer DNA extraction kit too and three of which were also positive with phenol-chloroform. One sample was positive only by phenol-chloroform and four only using Bioneer kit. The 32 resting samples were negative by all three methods.
Various studies have reported different methods for DNA extraction. Hsu et al., extracted DNA from Legionella cells by freeze & thaw [14]. Different kits have also been used for DNA release from bacterial cells. Morio and co-workers applied a DNA Mini Kit (Qiagen) for extracting DNA in water samples to identify the presence of Legionella[1]. In another study, Ariefjohan and co-workers extracted total DNA from human fecal specimens utilizing four different kits [15]. They found a significant disparity in DNA extraction yield and concluded that extraction kits incorporated lysing matrix and vigorous shaking produced high quality DNA [15]. Tomaso and co-workers used five commercial kits for detection of Brucellae in tissue specimens. They observed significant differences in DNA yield as high as two orders of magnitude for some samples [23]. Other studies have also concluded that the extraction methods play an important role with regard to performance in downstream molecular applications [24, 25].
Excluding the freeze & thaw method, in the present study a meaningful relationship was found among other three methods in which DNA extraction using Bioneer kit and to a lesser degree phenol-chlorophorm methods showed higher sensitivities. No Legionella were detected by freeze & thaw method, though 8.9, 17.7, and 26.6 percent of samples yielded Legionella contamination when Qiagen kit, classic phenol-chlorophorm and Bioneer kit methods were used, respectively. Two assumptions can be made here: a) relatively low concentrations of Legionella were present in these samples; b) the depletion of DNA amplification capacity rooted from the composition of water samples and presence of PCR inhibitors such as calcium and magnesium ions which may interfere with DNA extraction reagents and cause inhibition. Insufficient DNA yield due to low concentrations of Legionella could have led to negative results in our study when the method of freeze & thaw was conducted. Moreover, DNA yield might be out of the range of conventional PCR assay because of the small amount of bacteria in these samples that reflected the situation under real anthropogenic environmental conditions. This also indicates that DNA purification may be more relevant and a critical step when the amount of DNA in sample is very low [20]. Higher DNA yield indeed increases recovery of DNA from the bacterial community in a sample [22] and thus enhances chances of detecting rare species, such as Legionella. Comparing classic manual protocol and commercial kits for determination of agreement rate in our study, by excluding the freeze and thaw method in the further analysis of DNA extraction methods, showed that the interrelate analysis are Kappa coefficient calculated as 0.619 with P < 0.001 (Table 3). This measure of agreement is statistically significant. Most statisticians prefer Kappa values to be at least 0.6 and most often higher than 0.7 before claiming a good level of agreement [26]. Similarly, comparing Bioneer kit to the classic phenol-chlorophorm manual method illustrated the same good agreement (Kappa = 0.619); however, a moderate agreement was observed comparing phenol-chlorophorm method with Qiagen kit (Kappa = 0.433). Comparison of two commercial kits for determination of agreement rate, the results of interrelate analysis indicated Kappa coefficient calculated as 0.423 with P < 0.001 (Table 3). This measure of agreement inferred a statistically moderate agreement. As a rule of thumb, values of Kappa between 0.41-0.6 are considered moderate agreement. It should be also noted that a reference test is generally used to determine the sensitivity and specificity of a test or an instrument such as a kit against another one, which was not the goal of present study.
In general, DNA extraction using columned Bioneer kit followed by phenol-chloroform found to be the most appropriate methods in this study as compared to others. Considering these findings, the use of a reliable, useful and adapted DNA extraction protocol in such samples is therefore of high importance.
Legionella prevalence and species identification
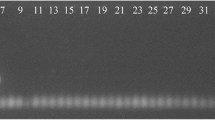
Identification of Legionella specie was based on the presence of an amplified product of 654 bp (Figure 1). Analysis of PCR results revealed a great diversity with regard to the sources from which samples were taken. Table 2 also demonstrates the results of Legionella monitoring in the seven hospitals by the source. In general, showerheads were the most contaminated source with 58.3 percent positive samples. Similarly, the samples from cooling waters, hot water taps, and hot water tanks yielded fairly comparable results (30.8%, 25%, and 25%, respectively). However, this was not the case with the cold-water tap samples and only 9% were positive for Legionella. For medical units with a high risk of legionellosis five positive samples in all were detected [Pediatrics stem cell transplantation (0), Bone marrow transplantation (0), Cancer Dep. (1), Maternity (1), Cardiac surgery (0), ENT (2), and Infant (1) wards].
Two positive samples were identified for species by DNA sequencing. DNA for sequencing was prepared by the 16S rRNA. The PCR products of two Legionella isolates were sequenced at MWG (Germany), DNA sequence was used to search the Gene Bank database, and the database entry with the highest percentage similarity was taken to identify the species. Nucleotide sequences data have been submitted to the Gene Bank database with accession Nos. FJ480932 and FJ480933.
Detection of Legionella in aquatic environments has been demonstrated in other researches [14, 27, 28]. Although PCR inhibitor may interfere with the results obtained by PCR, it has shown higher sensitivity than culture as demonstrated by Lye et al., and Morio et al., [1, 29]. This may be derived from the relatively low concentrations of Legionella and supported by the fact that Legionella bacteria are commonly present in aquatic environments in the viable but non-culturable status which cannot be detected by culture [30, 31]. In our study, although no statistical difference among Legionella positive rates in various sources was found (P > 0.05), the positive rate itself showed the severity of contamination. Therefore, even though the results obtained by PCR are not a valid determinant of Legionella viability in the environmental samples, it should be seriously considered as a potential public health threat. Owing to the lack of epidemiological and ecological studies, no Legionella outbreaks have been reported in environmental water samples in Tehran or other cities of Iran till now. However, considering patients’ complaint about their acquired pulmonary diseases at hospitals, the results of this study showed that sporadic or even a fairly high incidence of Legionella might have occurred but neglectfully distinguished as other pulmonary diseases.
Conclusion
Considering the important role of DNA extraction methods with regard to performance in downstream molecular applications and the usual low amount of bacteria in environmental water samples such as hospital water, the use of optimised methods in detection of Legionella by PCR assays is critical. Molecular techniques based on PCR assay offer a rapid, practical, cost-effective and sensitive alternative for detection of Legionella. Although the concentration of Legionellae in the sampled hospital water systems was not determined, given the high positive rate of Legionella colonization, hospital-acquired legionellosis might be under diagnosed in Tehran. It calls for urgent control measures to minimize the transmission rate of Legionella from the source to the host and to prevent an outbreak.
References
Morio F, Corvec S, Caroff N, Legallou F, Drugeon H, Reynaud A: Real-time PCR assay for the detection and quantification of Legionella pneumophila in environmental water samples: utility for daily practice. Int J Hyg Environ Health 2008, 211(3–4):403–411.
Leont E, Legnani P, Bucci Sabattini M, Righi F: Prevalence of Legionella spp. in swimming pool environment. Water Res 2001, 35(15):3749–3753. 10.1016/S0043-1354(01)00075-6
Sambrook J, Russell DW: Molecular Cloning: a Laboratory Manual. 3rd edition. Cold Spring Harbor, New York: CSHL press; 2001.
Miyamoto H, Yamamoto H, Arima K, Fujii J, Maruta K, Izu K, Shiomori T: Development of a new seminested PCR method for detection of Legionella species and its application to surveillance of Legionella in hospital cooling tower water. Appl Environ Microbiol 1997, 63(7):2489–2494.
Fields BS, Benson RF, Besser RE: Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev 2002, 15: 506–526. 10.1128/CMR.15.3.506-526.2002
Doust RH, Mobarez AM, Esmailli D: Detection of Legionella in hospital water supply using mip based primers. Int J Biol Sci 2008, 8: 930–934. 10.3923/jbs.2008.930.934
CDC: Legionellosis—United States, 2000–2009. MMWR Morb Mortal Wkly Rep 2011, 60(32):1083–1086. http://www.cdc.gov/Legionella/index.html
Joseph CA, Ricketts KD, Yadav R, Patel S, European Working Group for Legionella Infections: Travel-associated Legionnaires’ disease in Europe in 2009. Euro Surveill 2010, 15(41):19683.
The CDC investigation of Legionnaires’ disease among patients at the VA Pittsburgh Healthcare System 2013.
Joly P, Falconnet PA, Andre J, Weill N, Reyrolle M, Vanendesch F, Maurin M, Etienne J, Jarraud S: Quantitative real-time Legionella PCR for environmental water samples: data interpretation. Appl Environ Microbiol 2006, 72(4):2801–2808. 10.1128/AEM.72.4.2801-2808.2006
Yu VL: Cooling towers and legionellosis: A conundrum with proposed solutions. Int J Hyg Environ Health 2008, 211: 229–234. 10.1016/j.ijheh.2008.02.003
Lee JV, West AA: Survival and growth of Legionella species in the environment. J Appl Microbiol Symp Suppl 1991, 70: 121–129.
Bartie C, Venter SN, Nel LH: Identification methods for Legionella from environmental samples. Water Res 2003, 37(6):1362–1370. 10.1016/S0043-1354(02)00220-8
Hsu BM, Chen CH, Wan MT, Cheng HW: Legionella prevalence in hot spring recreation areas of Taiwan. Water Res 2006, 40(17):3267–3273. 10.1016/j.watres.2006.07.007
Arief Johan MW, Savaiano DA, Nacatsu CH: Comparison of DNA extraction kits for PCR-DGGE analysis of human intestinal microbial communities from fecal specimens. Nutr J 2010, 9: 23–28. 10.1186/1475-2891-9-23
Murphy MA, Shariflou MR, Moran C: High quality genomic DNA extraction from large milk samples. J Dairy Res 2009, 69: 645–649.
Lopez-Calleja I, Gonzalez I, Fajardo V, RodrIguez M, Hernandez P, Garcia T, Martin R: Rapid detection of cows' milk in sheeps' and goats' milk by a species-specific polymerase chain reaction technique. J Dairy Sci 2004, 87(9):2839–2840. 10.3168/jds.S0022-0302(04)73412-8
Pinto AD, Forte VT, Guastadisegni MC, Martin OC, Schena FP, Tanttilo G: A comparison of DNA extraction methods for food analysis. Food control 2007, 18(1):76–80. 10.1016/j.foodcont.2005.08.011
Whitehouse CA, Hottel HE: Comparison of five commercial DNA extraction kits for the recovery of Francisella tularensis DNA from spiked soil samples. Mol Cell Protemics 2007, 21: 92–96. 10.1016/j.mcp.2006.08.003
Queipo-Ortuño M, Tena F, Colmenero J, Morata P: Comparison of seven commercial DNA extraction kits for the recovery of Brucella DNA from spiked human serum samples using real-time PCR. Eur J Clin Microbiol Infect D 2008, 27(2):109–114. 10.1007/s10096-007-0409-y
Dauphin LA, Moser BD, Bowen MD: Evaluation of five commercial nucleic acid extraction kits for their ability to inactivate Bacillus anthracis spores and comparison of DNA yields from spores and spiked environmental samples. J Microbiol Meth 2009, 76: 30–37. 10.1016/j.mimet.2008.09.004
Dauphin L, Stephens K, Eufinger S, Bowen M: Comparison of five commercial DNA extraction kits for the recovery of Yersinia pestis DNA from bacterial suspensions and spiked environmental samples. Appl Environ Microbiol 2010, 108: 163–172. 10.1111/j.1365-2672.2009.04404.x
Tomaso H, Kattar M, Eickhoff M, Wernery U, Al Dahouk S, Straube E, Neubauer H, Scholz H: Comparison of commercial DNA preparation kits for the detection of Brucellae in tissue using quantitative real-time PCR. BMC Infect Dis 2010, 10(100):1–5.
Cornelis H, Jan D, Vander Linden Johannes SP, Mirjam H: Comparative analysis of four methods to extract DNA from paraffin-embedded tissues: effect on downstream molecular applications. BMC Res Note 2010, 14(3):239–247.
Scupham A, Jones J, Wesley I: Comparison of DNA extraction methods for analysis of turkey fecal microbiota. J Appl Microbiol 2007, 102(2):401–409.
Landis JR, Koch GG: The measurement of observer agreement for categorical data. Biometrics 1997, 33(1):159–174.
Miller LA, Beebe JL, Butler JC, Martin W, Benson R, Hoffman RE, Fields BS: Use of polymerase chain reaction in an epidemiologic investigation of Pontiac fever. J Infect Dis 1993, 168(3):769–772. 10.1093/infdis/168.3.769
Roll BM, Fujioka RS: Detection of Legionella bacteria in sewage by polymerase chain reaction and standard culture method. Water Sci Technol 1995, 31: 409–416.
Lye D, Fout GS, Crout SR, Danielson R, Thio CL, Paszko-Kolva CM: Survey of ground, surface, and potable waters for the presence of Legionella species by EnviroAmpR PCR Legionella Kit, culture, and immunofluorescent staining. Water Res 1997, 31(2):287–293. 10.1016/S0043-1354(96)00252-7
Steinert M, Emody L, Amann R, Hacker J: Resuscitation of viable but nonculturable Legionella pneumophila Philadelphia JR32 by Acanthamoeba castellanii. Appl Environ Microbiol 1997, 63(5):2047–2053.
Hay J, Seal DV, Billcliffe B, Freer JH: Non-culturable Legionella pneumophila associated with Acanthamoeba castellanii: detection of the bacterium using DNA amplification and hybridization. J Appl Bacteriol 1995, 78: 61–65. 10.1111/j.1365-2672.1995.tb01674.x
Acknowledgements
This study was funded by Tehran University of Medical Sciences, Department of Environmental Health Engineering. The authors would like to thank Dr. A. Mohabati Mobarez for providing positive samples and the hospitals’ staff for their collaboration in this research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MR, HH and AM participated in design of the study and performed experimental and molecular genetic studies. MH participated in statistical analysis. MR and MJ drafted the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Rafiee, M., Jahangiri-rad, M., Hajjaran, H. et al. Detection and identification of Legionella species in hospital water supplies through Polymerase Chain Reaction (16S rRNA). J Environ Health Sci Engineer 12, 83 (2014). https://doi.org/10.1186/2052-336X-12-83
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2052-336X-12-83