Effect of a Stannous Fluoride Dentifrice on Biofilm Composition, Gene Expression and Biomechanical Properties
Abstract
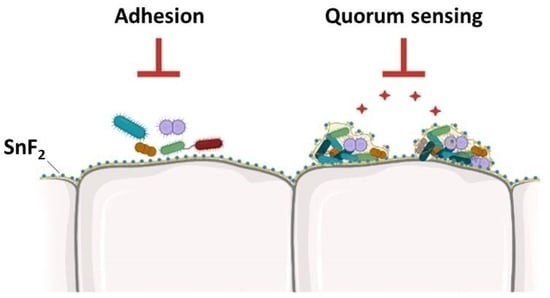
:1. Introduction
2. Materials and Methods
2.1. Inclusion Criteria
2.2. Exclusion Criteria
2.3. Study Visits and Clinical Procedures
2.4. In Situ Model (Retainer with HAP Discs) Preparation and Design
2.5. Intra-Oral Appliance Wear Instructions
2.6. Clinical Study Design
2.7. Confocal Laser-Scanning Microscopy (LSM)
2.8. Atomic Force Microscopy (AFM)
2.9. DNA Extraction and 16S Amplicon Sequencing
2.10. Bioinformatic Analysis for 16S rRNA Gene Amplicon Sequencing
2.11. Metatranscriptomics
3. Results and Discussion
3.1. SnF2 Toothpaste Affects Biofilm Growth and Morphology
3.2. SnF2 Toothpaste Alters Nanostructure and Biomechanical Properties of the Biofilm
3.3. SnF2 Toothpaste Maintains the Natural Healthy Oral Microflora
3.4. Gene Expression of Supragingival Plaque
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eke, P.I.; Thornton-Evans, G.O.; Wei, L.; Borgnakke, W.S.; Dye, B.A.; Genco, R.J. Periodontitis in US Adults: National Health and Nutrition Examination Survey 2009–2014. J. Am. Dent. Assoc. 2018, 149, 576–588. [Google Scholar] [CrossRef]
- Könönen, E.; Gursoy, M.; Gursoy, U.K. Periodontitis: A Multifaceted Disease of Tooth-Supporting Tissues. J. Clin. Med. 2019, 8, 1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, J.D.; Papapanou, P.N.; Philips, K.H.; Offenbacher, S. Periodontal Medicine: 100 Years of Progress. J. Dent. Res. 2019, 98, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Schoen, M.H.; Freed, J.R. Prevention of dental disease: Caries and periodontal disease. Annu. Rev. Public Health 1981, 2, 71–92. [Google Scholar] [CrossRef]
- Koo, H.; Yamada, K.M. Dynamic cell-matrix interactions modulate microbial biofilm and tissue 3D microenvironments. Curr. Opin Cell. Biol. 2016, 42, 102–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm Matrixome: Extracellular Components in Structured Microbial Communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Abebe, G.M. Oral Biofilm and Its Impact on Oral Health, Psychological and Social Interaction. Int. J. Oral Dent. Health 2021, 7, 127–137. [Google Scholar]
- Ten Cate, J.M.; Buzalaf, M.A.R. Fluoride Mode of Action: Once There Was an Observant Dentist. J. Dent. Res. 2019, 98, 725–730. [Google Scholar] [CrossRef]
- Sanz, M.; Serrano, J.; Iniesta, M.; Santa Cruz, I.; Herrera, D. Antiplaque and antigingivitis toothpastes. Monogr. Oral Sci. 2013, 23, 27–44. [Google Scholar]
- Serrano, J.; Escribano, M.; Roldán, S.; Martín, C.; Herrera, D. Efficacy of adjunctive anti-plaque chemical agents in managing gingivitis: A systematic review and meta-analysis. J. Clin. Periodontol. 2015, 42 (Suppl. 16), S106–S138. [Google Scholar] [CrossRef]
- Rajendiran, M.; Trivedi, H.M.; Chen, D.; Gajendrareddy, P.; Chen, L. Recent Development of Active Ingredients in Mouthwashes and Toothpastes for Periodontal Diseases. Molecules 2021, 26, 2001. [Google Scholar] [CrossRef] [PubMed]
- Haraszthy, V.I.; Raylae, C.C.; Sreenivasan, P.K. Antimicrobial effects of a stannous fluoride toothpaste in distinct oral microenvironments. J. Am. Dent. Assoc. 2019, 150, S14–S24. [Google Scholar] [CrossRef] [PubMed]
- Konradsson, K.; Lingström, P.E.; Emilson, C.G.; Johannsen, G.; Ramberg, P.; Johannsen, A. Stabilized stannous fluoride dentifrice in relation to dental caries, dental erosion and dentin hypersensitivity: A systematic review. Am. J. Dent. 2020, 33, 95–105. [Google Scholar] [PubMed]
- Trentin, G.A.; Mendes, L.T.; Silva, B.S.D.; Casagrande, L.; Araujo, F.B.D.; Lenzi, T.L. Reduction in erosive tooth wear using stannous fluoride-containing dentifrices: A meta-analysis. Braz. Oral Res. 2021, 35, 1–12. [Google Scholar] [CrossRef]
- Seriwatanachai, D.; Triratana, T.; Kraivaphan, P.; Amaornchat, C.; Mateo, L.R.; Sabharwal, A.; Delgado, E.; Szewczyk, G.; Ryan, M.; Zhang, Y.-P. Effect of stannous fluoride and zinc phosphate dentifrice on dental plaque and gingivitis: A randomized clinical trial with 6-month follow-up. J. Am. Dent. Assoc. 2019, 150, S25–S231. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Li, X.; Liu, H.; Mateo, L.R.; Sabharwal, A.; Xu, G.; Szewczyk, G.; Ryan, M.; Zhang, Y.-P. Evaluation of a stabilized stannous fluoride dentifrice on dental plaque and gingivitis in a randomized controlled trial with 6-month follow-up. J. Am. Dent. Assoc. 2019, 150, S32–S37. [Google Scholar] [CrossRef]
- Hines, D.; Xu, S.; Stranick, M.; Lavender, S.; Pilch, S.; Zhang, Y.; Sullivan, R.; Montesani, L.; Montesani, L.; Mateo, L.; et al. Effect of a stannous fluoride toothpaste on dentinal hypersensitivity: In vitro and clinical evaluation. J. Am. Dent. Assoc. 2019, 150, S47–S59. [Google Scholar] [CrossRef]
- Myers, C.P.; Pappas, I.; Makwana, E.; Begum-Gafur, R.; Utgikar, N.; Alsina, M.A.; Fitzgerald, M.; Trivedi, H.M.; Gaillard, J.F.; Masters, J.G.; et al. Solving the problem with stannous fluoride: Formulation, stabilization, and antimicrobial action. J. Am. Dent. Assoc. 2019, 150, S5–S13. [Google Scholar] [CrossRef]
- Cvikl, B.; Lussi, A.; Carvalho, T.S.; Moritz, A.; Gruber, R. Stannous chloride and stannous fluoride are inhibitors of matrix metalloproteinases. J. Dent. 2018, 78, 51–58. [Google Scholar] [CrossRef]
- Lussi, A.; Carvalho, T.S. The future of fluorides and other protective agents in erosion prevention. Caries Res. 2015, 49, 18–29. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Liu, J.; Li, J.; Zhou, X.; Wang, L.; Liu, J.; Xu, X. Comparative effect of a stannous fluoride toothpaste and a sodium fluoride toothpaste on a multispecies biofilm. Arch. Oral Biol. 2017, 74, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Duran-Pinedo, A.E. Metatranscriptomic analyses of the oral microbiome. Periodontology 2000, 85, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Sedghi, L.; Ganther, S.; Malone, E.; Kamarajan, P.; Kapila, Y.L. Host-microbe interactions: Profiles in the transcriptome, the proteome, and the metabolome. Periodontology 2020, 82, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Lavender, S.; Woo, J.; Guo, L.; Shi, W.; Kilpatrick-Liverman, L.; Gimzewski, J. Nanoscale characterization of effect of L-arginine on streptococcus mutans biofilm adhesion by atomic force microscopy. Microbiology 2014, 160, 1466–1473. [Google Scholar] [CrossRef] [PubMed]
- Kundukad, B.; Seviour, T.; Liang, Y.; Rice, S.A.; Kjelleberg, S.; Doyle, P.S. Mechanical properties of the superficial biofilm layer determine the architecture of biofilms. Soft Matter 2016, 12, 5718–5726. [Google Scholar] [CrossRef] [Green Version]
- Baniasadi, M.; Xu, Z.; Gandee, L.; Du, Y.; Zimmern, P.; Minary-Jolandan, M. Nanoindentation of Pseudomonas aeruginosa bacterial biofilm using atomic force microscopy. Mater. Res. Express 2014, 1, 045411. [Google Scholar] [CrossRef]
- Zhu, B.; Macleod, L.C.; Newsome, E.; Liu, J.; Xu, P. Aggregatibacter actinomycetemcomitans mediates protection of Porphyromonas gingivalis from Streptococcus sanguinis hydrogen peroxide production in multi-species biofilms. Sci. Rep. 2019, 9, 4944. [Google Scholar] [CrossRef] [Green Version]
- Al-Ahmad, A.; Roth, D.; Wolkewitz, M.; Wiedmann-Al-Ahmad, M.; Follo, M.; Ratka-Krüger, P.; Deimling, D.; Hellwig, E.; Hannig, C. Change in diet and oral hygiene over an 8-week period: Effects on oral health and oral biofilm. Clin. Oral Investig. 2010, 14, 391–396. [Google Scholar] [CrossRef]
- Wake, N.; Asahi, Y.; Noiri, Y.; Hayashi, M.; Motooka, D.; Nakamura, S.; Gotoh, K.; Miura, J.; Machi, H.; Iida, T.; et al. Temporal dynamics of bacterial microbiota in the human oral cavity determined using an in situ model of dental biofilms. npj Biofilms Microbiomes 2016, 2, 16018. [Google Scholar] [CrossRef]
- Krishnan, K.; Chen, T.; Paster, B.J. A practical guide to the oral microbiome and its relation to health and disease. Oral Dis. 2017, 23, 276–286. [Google Scholar] [CrossRef] [Green Version]
- Thomas, G.; Burnham, N.A.; Camesano, T.A.; Wen, Q. Measuring the mechanical properties of living cells using atomic force microscopy. J. Vis. Exp. 2013, 76, 50497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asally, M.; Kittisopikul, M.; Rué, P.; Du, Y.; Hu, Z.; Cagatay, T.; Robinson, A.B.; Lu, H.; García-Ojalvo, J.; Süel, G.M. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc. Natl. Acad. Sci. USA 2012, 109, 18891–18896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Studio Team. RStudio: Integrated Development Environment for R. RStudio; PBC: Boston, MA, USA, 2021; Available online: http://www.rstudio.com/ (accessed on 4 July 2022).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqcFastQC (accessed on 4 July 2022).
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.-A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 33, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Escapa, I.F.; Huang, Y.; Chen, T.; Lin, M.; Kokaras, A.; Dewhirst, F.E.; Lemon, K.P. Construction of habitat-specific training sets to achieve species-level assignment in 16S rRNA gene datasets. Microbiome 2020, 15, 65. [Google Scholar] [CrossRef]
- Davis, N.M.; Proctor, D.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-7. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 4 July 2022).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 4 July 2022).
- Ram, K.; Wickham, H. Wesanderson: A Wes Anderson Palette Generator. R Package Version 0.3.6. 2018. Available online: https://CRAN.R-project.org/package=wesanderson (accessed on 4 July 2022).
- Wilke, C.O. Cowplot: Streamlined Plot Theme and Plot Annotations for ‘Ggplot2’. R Package Version 1.1.1. 2020. Available online: https://CRAN.R-project.org/package=cowplot (accessed on 4 July 2022).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, W.; Strunk, O.; Westram, R.; Richter, L.; Meier, H.; Yadhukumar, B.A.; Lai, T.; Steppi, S.; Jobb, G.; Förster, W.; et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004, 32, 1363–1371. [Google Scholar] [CrossRef] [Green Version]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup, The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.H.; Davis, F.G.; et al. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.; Papanicolaou, A. TransDecoder (Find Coding Regions within Transcripts) 2016. Available online: https://github.com/TransDecoder/TransDecoder (accessed on 4 July 2022).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [Green Version]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. EggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 4 July 2022).
- Tarazona, S.; García, F.; Ferrer, A.; Dopazo, J.; Conesa, A. NOIseq: A RNA-seq differential expression method robust for sequencing depth biases. EMBnet 2012, 17, 18–19. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Wei, T.; Simko, V. Corrplot: Visualization of a Correlation Matrix. 2021. (Version 0.92). Available online: https://github.com/taiyun/corrplot (accessed on 4 July 2022).
- Harrell, F., Jr. Hmisc: Harrell Miscellaneous_.R Package Version 4.6-0. 2021. Available online: https://CRAN.R-project.org/package=Hmisc (accessed on 4 July 2022).
- Le, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Bhatnagar, S. Interactive Q–Q and Manhattan plots using Plotly. Js. R Package Version 0.3.0. 2016. Available online: https://cran.r-project.org/web/packages/manhattanly (accessed on 4 July 2022).
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. R Package Version 1.10.0. 2021. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 4 July 2022).
- Slowikowski, K. Ggrepel: Automatically Position Non-Overlapping Text Labels with ‘ggplot2’. R package version 0.9.1. 2021. Available online: https://CRAN.R-project.org/package=ggrepel (accessed on 4 July 2022).
- Kassambara, A. Ggpubr: ‘ggplot2’ Based Publication Ready Plots. R Package Version 0.4.0. 2020. Available online: https://CRAN.R-project.org/package=ggpubr (accessed on 4 July 2022).
- Hamilton, N.E.; Ferry, M. Ggtern: Ternary Diagrams Using ggplot2. J. Stat. Softw. 2018, 87, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Horikoshi, M.; Li, W. Ggfortify: Unified Interface to Visualize Statistical Result of Popular R Packages. R J. 2016, 8, 474–485. [Google Scholar] [CrossRef] [Green Version]
- Horikoshi, M.; Tang, Y. Ggfortify: Data Visualization Tools for Statistical Analysis Results. 2018. Available online: https://CRAN.R-project.org/package=ggfortify (accessed on 4 July 2022).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; D’Agostino McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Wickham, H. Tidyr: Tidy Messy Data. R Package Version 1.1.4. 2021. Available online: https://CRAN.R-project.org/package=tidyr (accessed on 4 July 2022).
- Wickham, H. Reshaping data with the reshape package. J. Stat. Softw. 2007, 21, 1–20. [Google Scholar] [CrossRef]
- Wickham, H. The Split-Apply-Combine Strategy for Data Analysis. J. Stat. Softw. 2011, 40, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Dowle, M.; Srinivasan, A. Data.Table: Extension of ‘Data.Frame’. R Package Version 1.14.2. 2021. Available online: https://CRAN.R-project.org/package=data.table (accessed on 4 July 2022).
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps; R Package Version 1.0. 12; CRAN. R-Project: 2019. Available online: https://rdrr.io/cran/pheatmap/ (accessed on 4 July 2022).
- Khosravi, Y.; Palmer, S.; Daep, C.A.; Sambanthamoorthy, K.; Kumar, P.; Dusane, D.H.; Stoodley, P.A. Commercial SnF2 toothpaste formulation reduces simulated human plaque biofilm in a dynamic typodont model. J. Appl. Microbiol. 2022, 1–12. [Google Scholar] [CrossRef]
- Otten, M.P.T.; Busscher, H.J.; van der Mei, H.C.; van Hoogmoed, C.G.; Abbas, F. Acute and substantive action of antimicrobial toothpastes and mouthrinses on oral biofilm in vitro. Eur. J. Oral Sci. 2011, 119, 151–155. [Google Scholar] [CrossRef] [Green Version]
- Kirsch, J.; Hannig, M.; Winkel, P.; Basche, S.; Leis, B.; Pütz, N.; Kensche, A.; Hannig, C. Influence of pure fluorides and stannous ions on the initial bacterial colonization in situ. Sci. Rep. 2019, 9, 18499–18512. [Google Scholar] [CrossRef] [Green Version]
- Al Safadi, R.; Abu-Ali, G.S.; Sloup, R.E.; Rudrik, J.T.; Waters, C.M.; Eaton, K.A.; Manning, S.D. Correlation between in vivo biofilm formation and virulence gene expression in Escherichia coli O104:H4. PLoS ONE 2012, 7, e41628. [Google Scholar] [CrossRef]
- Cross, S.E.; Kreth, J.; Zhu, L.; Qi, F.; Pelling, A.E.; Shi, W.; Gimzewski, J.K. Atomic force microscopy study of the structure-function relationships of the biofilm-forming bacterium Streptococcus mutans. Nanotechnology 2006, 17, S1–S7. [Google Scholar] [CrossRef]
- Zhu, L.; Kreth, J.; Cross, S.E.; Gimzewski, J.K.; Shi, W.; Qi, F. Functional characterization of cell-wall-associated protein WapA in Streptococcus mutans. Microbiology 2006, 152, 2395–2404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebeer, S.; Verhoeven, T.L.; Francius, G.; Schoofs, G.; Lambrichts, I.; Dufrêne, Y.; Vanderleyden, J.; De Keersmaecker, S.C. Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl. Environ. Microbiol. 2009, 75, 3554–3563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebellenger, L.; Verrez-Bagnis, V.; Passerini, D.; Delbarre-Ladrat, C. Comparative genomics reveals a widespread distribution of an exopolysaccharide biosynthesis gene cluster among Vibrionaceae. BMC Res. Notes 2018, 11, 102–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjasupattananan, S.; Lai, C.S.; Persson, G.R.; Pjetursson, B.E.; Lang, N.P. Effect of a stannous fluoride dentifrice on the sulcular microbiota: A prospective cohort study in subjects with various levels of periodontal inflammation. Oral Health Prev. Dent. 2005, 3, 263–272. [Google Scholar]
- Anderson, A.C.; Al-Ahmad, A.; Schlueter, N.; Frese, C.; Hellwig, E.; Binder, N. Influence of the Long-Term Use of Oral Hygiene Products Containing Stannous Ions on the Salivary Microbiome—A Randomized Controlled Trial. Sci. Rep. 2020, 10, 9546–9553. [Google Scholar] [CrossRef]
- Kruse, A.B.; Schlueter, N.; Kortmann, V.K.; Frese, C.; Anderson, A.; Wittmer, A.; Hellwig, E.; Vach, K.; Al-Ahmad, A. Long-Term Use of Oral Hygiene Products Containing Stannous and Fluoride Ions: Effect on Viable Salivary Bacteria. Antibiotics 2021, 10, 481. [Google Scholar] [CrossRef]
- Eraso, J.M.; Markillie, L.M.; Mitchell, H.D.; Taylor, R.C.; Orr, G.; Margolin, W. The highly conserved MraZ protein is a transcriptional regulator in Escherichia coli. J. Bacteriol. 2014, 196, 2053–2066. [Google Scholar] [CrossRef] [Green Version]
- White, M.; Hough-Neidig, A.; Khan, S.; Eswara, P. MraZ is a transcriptional inhibitor of cell division in Bacillus subtilis. bioRxiv 2022. [Google Scholar] [CrossRef]
- Rismondo, J.; Schulz, L.M. Not Just Transporters: Alternative Functions of ABC Transporters in Bacillus subtilis and Listeria monocytogenes. Microorganisms 2021, 9, 163. [Google Scholar] [CrossRef]
- Zhu, X.; Long, F.; Chen, Y.; Knøchel, S.; She, Q.; Shi, X. A putative ABC transporter is involved in negative regulation of biofilm formation by Listeria monocytogenes. Appl. Environ. Microbiol. 2008, 74, 7675–7683. [Google Scholar] [CrossRef] [Green Version]
- Benda, M.; Schulz, L.M.; Stülke, J.; Rismondo, J. Influence of the ABC Transporter YtrBCDEF of Bacillus subtilis on competence, biofilm formation and cell wall thickness. Front. Microbiol. 2021, 12, 587035. [Google Scholar] [CrossRef]
- Turner, R.D.; Hobbs, J.K.; Foster, S.J. Atomic Force Microscopy Analysis of Bacterial Cell Wall Peptidoglycan Architecture. Methods Mol. Biol. 2016, 1440, 3–9. [Google Scholar]
- Hugouvieux-Cotte-Pattat, N.; Condemine, G.; Shevchik, V.E. Bacterial pectate lyases. Environ. Microbiol. Rep. 2014, 6, 427–440. [Google Scholar] [CrossRef]
- Yamazaki, A.; Li, J.; Hutchins, W.C.; Wang, L.; Ma, J.; Ibekwe, A.M.; Yang, C.H. Commensal effect of pectate lyases secreted from Dickeya dadantii on proliferation of Escherichia coli O157:H7 EDL933 on lettuce leaves. Appl. Environ. Microbiol. 2011, 77, 156–162. [Google Scholar] [CrossRef] [Green Version]
- Rice, K.C.; Bayles, K.W. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 2008, 72, 85–109. [Google Scholar] [CrossRef] [Green Version]
- Campoccia, D.; Montanaro, L.; Arciola, C.R. Extracellular DNA (eDNA). A Major Ubiquitous Element of the Bacterial Biofilm Architecture. Int. J. Mol. Sci. 2021, 22, 9100. [Google Scholar] [CrossRef]
- Gutierrez, J.A.; Crowder, T.; Rinaldo-Matthis, A.; Ho, M.C.; Almo, S.C.; Schramm, V.L. Transition state analogs of 5’-methylthioadenosine nucleosidase disrupt quorum sensing. Nat. Chem. Biol. 2009, 5, 251–257. [Google Scholar] [CrossRef]
- Paluch, E.; Rewak-Soroczyńska, J.; Jędrusik, I.; Mazurkiewicz, E.; Jermakow, K. Prevention of biofilm formation by quorum quenching. Appl. Microbiol. Biotechnol. 2020, 104, 1871–1881. [Google Scholar] [CrossRef] [Green Version]
- Wong, S.; Dessen, A. Structure of a bacterial α2-macroglobulin reveals mimicry of eukaryotic innate immunity. Nat. Commun. 2014, 5, 4917. [Google Scholar] [CrossRef] [Green Version]
- Schulz, A.; Schumann, W. HrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J. Bacteriol. 1996, 178, 1088–1093. [Google Scholar] [CrossRef] [Green Version]
- Roncarati, D.; Pinatel, E.; Fiore, E.; Peano, C.; Loibman, S.; Scarlato, V. Helicobacter pylori Stress-Response: Definition of the HrcA Regulon. Microorganisms 2019, 7, 436. [Google Scholar] [CrossRef] [Green Version]
- Hathroubi, S.; Zerebinski, J.; Ottemann, K.M. Helicobacter pylori biofilm involves a multigene stress-biased response, including a structural role for flagella. mBio 2018, 9, e01973-18. [Google Scholar] [CrossRef] [Green Version]
- van der Veen, S.; Abee, T. HrcA and DnaK are important for static and continuous-flow biofilm formation and disinfectant resistance in Listeria monocytogenes. Microbiology 2010, 156, 3782–3790. [Google Scholar] [CrossRef] [Green Version]
- Zenobia, C.; Darveau, R.P. Does Oral Endotoxin Contribute to Systemic Inflammation? Front. Oral Health 2022, 3, 911420. [Google Scholar] [CrossRef]
- Haught, C.; Xie, S.; Circello, B.; Tansky, C.S.; Khambe, D.; Klukowska, M.; Huggins, T.; White, D.J. Lipopolysaccharide and Lipoteichoic Acid Virulence Deactivation by Stannous Fluoride. J. Clin. Dent. 2016, 27, 84–89. [Google Scholar]
- Xie, S.; Haught, J.C.; Tansky, C.S.; Klukowska, M.; Hu, P.; Ramsey, D.L.; Circello, B.; Huggins, T.G.; White, D.J. Clinical effects of stannous fluoride dentifrice in reducing plaque microbial virulence III: Lipopolysaccharide and TLR2 reporter cell gene activation. Am. J. Dent. 2018, 31, 215–224. [Google Scholar]
- Green, E.R.; Mecsas, J. Bacterial Secretion Systems: An Overview. Microbiol. Spectr. 2016, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Paju, S.; Scannapieco, F.A. Oral biofilms, periodontitis, and pulmonary infections. Oral Dis. 2007, 13, 508–512. [Google Scholar] [CrossRef] [Green Version]
- Blaizot, A.; Vergnes, J.N.; Nuwwareh, S.; Amar, J.; Sixou, M. Periodontal diseases and cardiovascular events: Meta-analysis of observational studies. Int. Dent. J. 2009, 59, 197–209. [Google Scholar]
- Nakamura, Y.; Tagusari, O.; Saito, K.; Oyamada, S.; Honda, K.; Homma, N.; Miyamoto, R.; Nakano, K. Prevalence of dental disease in patients undergoing heart valve surgery. Jap. J. Cardiovasc. Surg. 2008, 37, 213–216. [Google Scholar] [CrossRef]












| Group | Sodium Monofluorophosphate | SnF2 |
|---|---|---|
| No. of reads (mean) | 199,766 | 214,510 |
| No. filtered reads (mean) | 146,682 | 155,574 |
| Total ASVs | 1034 | 1026 |
| Unique ASVs | 450 | 442 |
| ASVs ≥ 10 | 692 | 712 |
| Mean ASVs | 186 | 185 |
| ±sd | ±54.6 | ±59.9 |
| Shannon diversity index | 2.91 | 2.97 |
| ±sd | ±0.548 | ±0.437 |
| Simpson’s diversity index | 0.842 | 0.861 |
| ±sd | ±0.107 | ±0.069 |
| Taxa ID | Phylum | Genus Species | log2 FC | p-Value |
|---|---|---|---|---|
| ASV11 | Firmicutes | Veillonella parvula | 1.427 | 0.039 * |
| ASV13 | Proteobacteria | Neisseria flavescens | −1.976 | 0.054 |
| ASV45 | Absconditabacteria (SR1) | Absconditabacteria (SR1) [G-1] | −3.471 | 0.060 |
| ASV50 | Firmicutes | Streptococcus sanguinis | 1.267 | 0.097 |
| ASV73 | Firmicutes | Streptococcus sp. | 1.296 | 0.093 |
| ASV98 | Firmicutes | Veillonella sp. | −3.435 | 0.048 * |
| ASV106 | Proteobacteria | Haemophilus sp. | −5.845 | 0.047 * |
| ASV118 | Bacteroidetes | Prevotella sp. | −5.882 | 0.046 * |
| ASV122 | Firmicutes | Gemella sp. | −5.538 | 0.060 |
| ASV125 | Bacteroidetes | Alloprevotella sp. HMT914 | −5.755 | 0.051 |
| ASV126 | Bacteroidetes | Prevotella aurantiaca | −5.472 | 0.063 |
| ASV128 | Firmicutes | Parvimonas sp. | −3.147 | 0.073 |
| ASV185 | Firmicutes | Gemella sp. | −4.961 | 0.092 |
| ASV190 | Firmicutes | Veillonella sp. | 4.944 | 0.093 |
| ASV204 | Bacteroidetes | Bergeyella sp. | 5.039 | 0.087 |
| ASV235 | Bacteroidetes | Prevotella sp. | −5.760 | 0.050 |
| ASV242 | Actinobacteria | Actinomyces sp. | 5.742 | 0.051 |
| ASV325 | Fusobacteria | Leptotrichia sp. | −5.749 | 0.051 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gumber, H.K.; Louyakis, A.S.; Sarma, T.; Fabijanic, K.I.; Paul, R.; Mellenbruch, K.; Kilpatrick-Liverman, L. Effect of a Stannous Fluoride Dentifrice on Biofilm Composition, Gene Expression and Biomechanical Properties. Microorganisms 2022, 10, 1691. https://doi.org/10.3390/microorganisms10091691
Gumber HK, Louyakis AS, Sarma T, Fabijanic KI, Paul R, Mellenbruch K, Kilpatrick-Liverman L. Effect of a Stannous Fluoride Dentifrice on Biofilm Composition, Gene Expression and Biomechanical Properties. Microorganisms. 2022; 10(9):1691. https://doi.org/10.3390/microorganisms10091691
Chicago/Turabian StyleGumber, Hardeep Kaur, Artemis S. Louyakis, Tulika Sarma, Kristina Ivana Fabijanic, Reeba Paul, Kristen Mellenbruch, and Latonya Kilpatrick-Liverman. 2022. "Effect of a Stannous Fluoride Dentifrice on Biofilm Composition, Gene Expression and Biomechanical Properties" Microorganisms 10, no. 9: 1691. https://doi.org/10.3390/microorganisms10091691